Classify particles corresponding to each reference projection
verify-class-byview.spi reads: views/prj###/sel_proj , views/prj###/sel_part_byv and views/prj###/stkfilt .
The procedure classifies the particles corresponding to each reference projection. The idea is that good particles will fall into discrete classes, and non-particles will fall into different classes. Notable outputs include:| ¤ | views/prj###/sel_part_byclass_???: | Doc file listing particles belonging to a particular class. |
| ¤ | views/prj###/listclasses: | Doc file listing classes and average CCC value of particles belonging to that class. |
| ¤ | views/prj###/classavg???: | Average image of particles belonging to a particular class. |
Verify particles
For each each reference projection, the aligned particles will have been assigned to classes, and sorted by cross-correlation coefficient. Ideally, the good particles will fall into discrete classes, and non-particles into different discrete classes. Outliers can be added or removed individually.
Different levels of control over the particles kept are offered. Recommended is to open each montage of particles belonging to each class, and select good particles. The fastest method is to select entire classes of particles. In between the two, in terms of control, would be to manually select only the particles with the highest CCC values.
There are two display options:- Use:
verifybyview.py --
Documentation.
The simplest way to start it would be to, from the Averages/ directory, type:
verifybyview
Adjust the parameters in the initial popup window, click Continue, and a montage of class windows will appear. Prior installation of SPIRE is required. - WEB -- Documentation
Whichever display option chosen will read: views/prj###/listclasses , views/prj###/sel_part_byclass_???, views/prj###/classavg??? , & views/prj###/avgfilt . It creates:
| ¤ | views/prj###/sel_class_good: | List of classes containing good particles. |
| ¤ | views/prj###/sel_part_byhand###: | For each class, a list of good particles. |
Verify particles in each of the reference view and don't forget to save views/prj###/sel_class_good.
Combine particles from good classes
verify-combine-classes.spi reads: views/prj###/sel_proj , views/prj###/sel_class_good , views/prj###/sel_part_byv , views/prj###/sel_part_byclass_???, & views/prj###/sel_part_byhandIt collects and formats the information in the form of prj###/sel_part_byclass. It creates:
| ¤ | views/prj###/sel_part_byv_good: | For each view, a list of selected particles. |
| ¤ | views/prj###/sel_part_byv_bad: | For each view, a list of deselected particles. |
| ¤ | views/parts_vsview_good: | Summary of particles kept for each view. |
[Optional] Re-screen the particles by view
For an additional layer of quality control, the newly created lists of good and bad particles by class can be re-screened for outliers.
To look for bad particles in the selection file, in the directory: Averages/Views,
type (with the appropriate extension):
montagefromdoc.py prj001/sel_part_byv_bad.dat prj001/avgfilt.dat
There should be a settings file, .montagefromdoc,
present with reasonable settings.
Select only the bad particles, and write the selection file to:
views/prj###/sel_part_byv_notgood.
Conversely, to look for good particles among the de-selected particles,
in the directory:Averages/Views, type (with the appropriate extension):
montagefromdoc.py prj001/sel_part_byv_good.dat prj001/avgfilt.dat
Select only the good particles, and write the selection to:
views/prj###/sel_part_byv_notbad.
Both of the above two steps are optional, i.e., you can perform one or the other, or both. If you run either one, you'll need to run the following procedure.
To update the lists of particles, run verify-recheck.spi. Among other files it reads: views/prj###/sel_part_byv_good, views/prj###/sel_part_byv_bad , views/prj###/sel_part_byv_notbad [optional], and views/prj###/sel_part_byv_notgood [optional]. It creates:| ¤ | views/prj###/sel_part_byv_goodB: | An updated list of selected particles, for each view. |
| ¤ | views/prj###/sel_part_byv_badB: | An updated list of deselected particles, for each view. |
| ¤ | summary_recheckB: | Summary of particles kept for each view. |
If this step is performed, remember in later steps to substitute views/prj###/sel_part_byv_goodB where appropriate.
[Optional] Check the spatial distribution of views (angular directions) for the reconstruction
The following two procedures are same as those run previously, except for one input setting used. Namely, change the parameter [wantGoodYN] to 1 to use the verified particles.
plot-ref-views.spi reads:
views/parts_vsview_good.
It creates a script file with gnuplot commands:
| ¤ | parts_vsview_good.gnu | Gnuplot script to plot histogram of number of particles vs projection view. |
|
The output Gnuplot script parts_vsview_good.gnu will plot number of particles vs view after verification. The histogram plot should be displayed automatically, if not click here.
|
| |
The output Gnuplot script parts_vsview_good.gnu will plot number of particles vs viewafter verification. The histogram plot should be displayed automatically, if not click here.
show-ref-views.spi reads: ../Alignment/refangles, and views/part_vsview_good. Change the parameter [wantGoodYN] to 1 to use the verified particles. It creates:| ¤ | show_ref_views_good: | SPIDER image showing distribution of sample images among the various reference projections. |
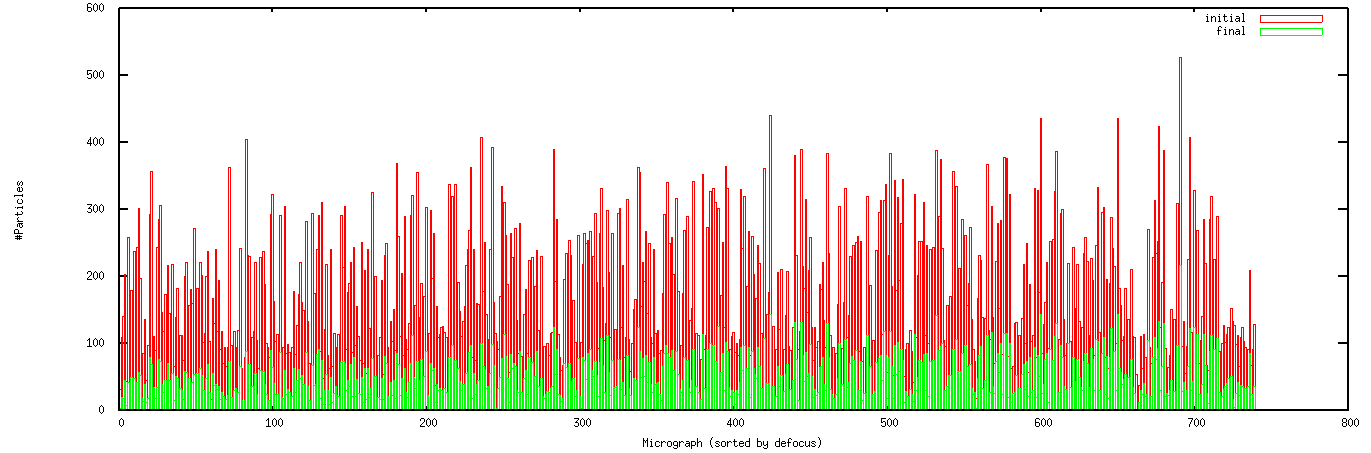
[Optional] Compare number of particles before and after verification as a function of defocus level.
With micrographs from all defocus values combined, it is possible that close-to-focus images may be discriminated against, for example because of lower CC values or simply being harder to see.
parts-vs-defocus.spi sorts the micrographs by estimated defocus and tabulates number of particles retained. It reads: ../Power_Spectra/defocus , sel_part_combined_good , ../Particles/win/glonum2micwin , and creates:| ¤ | Micrographs/sel_part_bymic_****: | Updated list of selected particles, for each micrograph. |
| ¤ | parts_vsmic: | Summary file listing the number of particles before and after verification. |
| ¤ | parts_vsdefocus.gnu: | Gnuplot script for plotting particles kept as a function of defocus. |
|
The output Gnuplot script good_parts_vsmic.gnu
will overlay the number of particles before and after verification.
The histogram plot should be displayed
automatically, if not click here.
See example on right: |
 |
References |
- Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J. (2008) Particle-verification for single-particle, reference-based-reconstruction using multivariate data analysis and classification. Journal of Structural Biology, Volume 164, Pages 41-48.
Page updated: 2016/10/19 Author: Tapu Shaikh