(modified 2009 oct 31)
montagefromdoc.py usage
Screen captures may not be up-to-date
Rationale:
Often, I would like to display more images than fits on a screen. JWEB is inconvenient, for several reasons. montage-spican open subsets of images, but it is not straightforward to manually page through screenfuls of images. The ability to page through screenfuls was the main feature I wanted to implement.
Getting started:
The SPIRE (SPIDER's Reconstruction Engine) package must be installed if not already present.
Procedure:
Assuming the Python script is in your $PATH, run it by typing:
montagefromdoc &
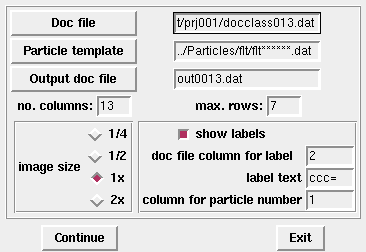
If you're running it for the first time, a pop-up window will appear with some pre-set defaults. When you change or update the filenames, they get saved. In the following example, I've picked up from a previous session, and the programs recalls those filenames:

If your data extension is not .dat, be sure to change it here.
In the example above, I had been using unstacked images but montagefromdoc also works with stacked images:
It may be preferable to enter the input file templates on the command line. (Navigating through the file-browser button can be cumbersome.) The first optional cammand-line argument is the doc file:
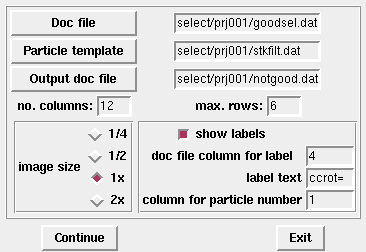
montagefromdoc select/prj001/goodsel.dat &
The second optional cammand-line argument is the image stack/template:
montagefromdoc select/prj001/goodsel.dat select/prj001/stkfilt.dat &
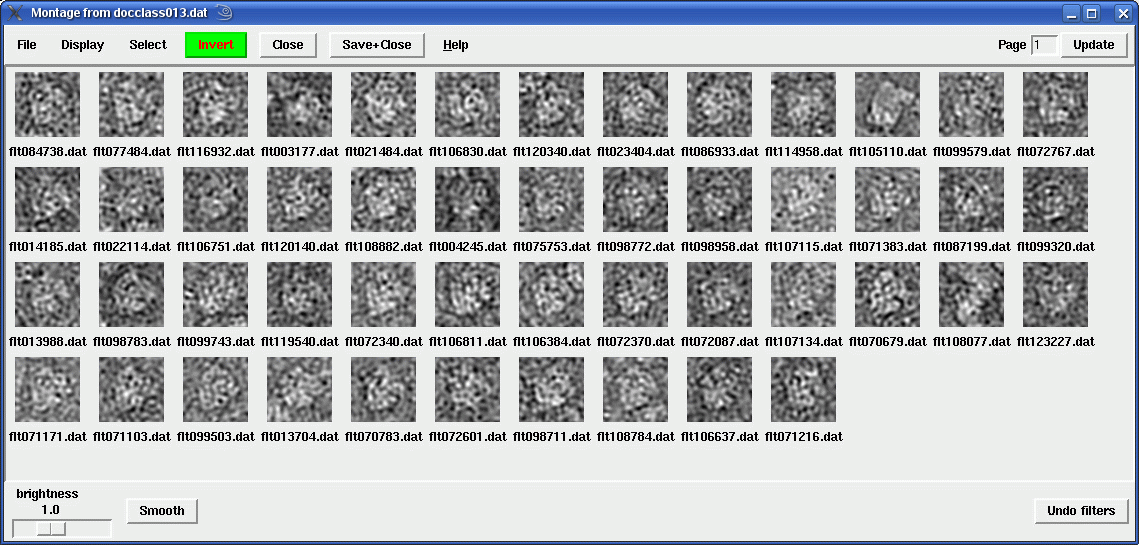
When you click on Continue, a montage of images will pop up:
With the menus overlain:
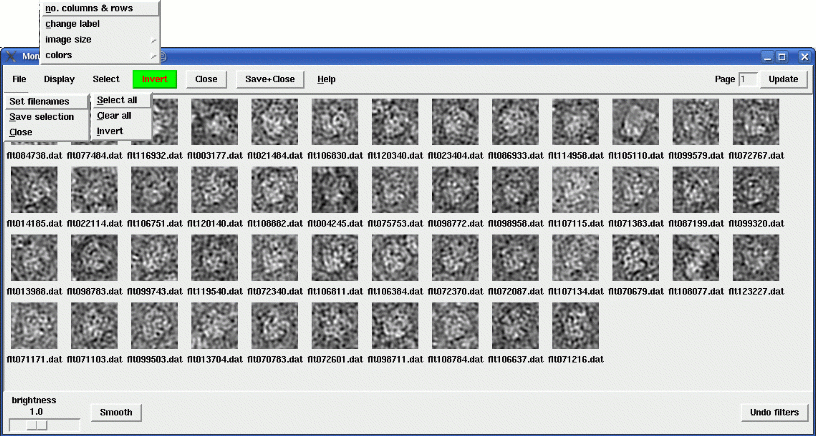
The selection part should be intuitive. Sometimes, clicking on the bad particles and inverting the selection is easier. Shift-clicking will select the contiguous stretch of particles from the last one you selected.
There are some keyboard shortcuts here (also available in Help-> Keyboard shortcuts:
From the Open Selections (CTRL-o) menu:
Modifications: